Familial clustering of myelodysplastic syndromes (MDSs) and acute myeloid leukemia (AML) can be caused by inherited factors. We screened 59 individuals from 17 families with 2 or more biological relatives with MDS/AML for variants in 12 genes with established roles in predisposition to MDS/AML, and identified a pathogenic germ line variant in 5 families (29%).
Known pathogenic germ line variants in 12 genes can explain nearly 30% of families with inherited predisposition to MDS/AML.
Asymptomatic carriers of germ line RUNX1 mutations develop detectable clonal hematopoiesis with a cumulative risk of >80% by age 50 years.
Abstract
Extending the screen with a panel of 264 genes that are recurrently mutated in de novo AML, we identified rare, nonsynonymous germ line variants in 4 genes, each segregating with MDS/AML in 2 families. Somatic mutations are required for progression to MDS/AML in these familial cases. Using a combination of targeted and exome sequencing of tumor and matched normal samples from 26 familial MDS/AML cases and asymptomatic carriers, we identified recurrent frameshift mutations in the cohesin-associated factor PDS5B, co-occurrence of somatic ASXL1 mutations with germ line GATA2 mutations, and recurrent mutations in other known MDS/AML drivers.
Mutations in genes that are recurrently mutated in de novo AML were underrepresented in the familial MDS/AML cases, although the total number of somatic mutations per exome was the same. Lastly, clonal skewing of hematopoiesis was detected in 67% of young, asymptomatic RUNX1 carriers, providing a potential biomarker that could be used for surveillance in these high-risk families.
Introduction
Myelodysplastic syndromes (MDSs) and acute myeloid leukemia (AML) are usually sporadic late-onset cancers, diagnosed at a median age >70 years. Rarely, MDS/AML has early onset and/or aggregates within families, suggesting inherited predisposition. Familial MDS/AML can occur in the context of syndromic bone marrow failure (eg, dyskeratosis congenita, severe congenital neutropenia, Fanconi anemia) or as Mendelian disorders that have MDS/AML as the principal clinical feature.
The Mendelian disorders include familial platelet disorder with associated myeloid malignancy (FPDMM) caused by germ line mutations in RUNX1 (OMIM #601399)1,2; familial AML with CEBPA mutation (OMIM #116897)3-5; the GATA2-associated syndromes (Emberger syndrome; dendritic cell, monocyte, B-lymphocyte, and natural killer–lymphocyte deficiency; and monocytopenia with susceptibility to mycobacterial, fungal, and papillomavirus infection and myelodysplasia)6-9; and other syndromes associated with germ line mutations in ANKRD26, SRP72, DDX41, or ETV6.10-13
Known genetic factors are estimated to explain fewer than half of apparently familial MDS/AML cases, but this has not been formally tested. In addition, the allelic spectrum of causal variants in these genes is not yet known. Furthermore, the variable latency and incomplete penetrance characteristic of most of these syndromes suggest that the acquisition of additional somatic mutations is required for MDS/AML initiation. The landscape of somatic alterations acquired in familial MDS/AML has not been well characterized. Lastly, hematopoietic cells from asymptomatic individuals carrying known pathogenic germ line variants have not been examined to determine whether potentially deleterious somatic mutations accumulate prior to clinical diagnosis of MDS/AML. We used a combination of targeted and unbiased sequencing to address these gaps in our understanding of the germ line and somatic genetics of familial MDS/AML.
Methods
Study participants
Seventy-one individuals from 21 families were included in the study (see supplemental Figure 1, available on the Blood Web site). Seventeen of these families had 2 or more cases of MDS and/or AML in biological relatives within 3 degrees of relation, 2 had a single case of MDS/AML and acquired bone marrow failure in a sibling, and the remaining 2 families were known carriers of deleterious RUNX1 alleles. The clinical characteristics of families 1001, 1015, and 1016 have been reported previously
Patients and unaffected family members provided informed consent to participate in and provide samples for protocols approved by the human studies committee at Washington University, The University of Chicago, Johns Hopkins University, or the University of Minnesota. Nonmalignant (“normal”) tissue (skin biopsy sample or buccal swab) was collected from all subjects. Tumor samples (from bone marrow aspirates or peripheral blood) were obtained from subjects with MDS/AML, and peripheral blood samples were obtained from asymptomatic carriers of known pathogenic germ line alleles. Sequencing, analysis, and interpretation were performed at The Genome Institute (Washington University), under a protocol authorizing whole-genome sequencing and data sharing approved by the Washington University Human Research Protection Office.
Sequence production
Genomic DNA was fragmented and hybridized in solution to capture probes, eluted, and sequenced on the HiSeq2500 platform (Illumina, San Diego, CA), as previously described.18 Three probe sets were used for hybridization capture. First, a familial MDS/AML germ line panel was designed to target 3 Mendelian MDS/AML genes and 9 genes most commonly associated with congenital bone marrow failure syndromes and predisposition to MDS/AML (supplemental Table 1). Biotinylated probes covering coding exons in all known transcripts of these genes were designed and synthesized (Integrated DNA Technologies, Coralville, IA).
The second panel was designed to target all exons of the 264 recurrently mutated genes (RMGs) in de novo MDS/AML. This list includes all genes mutated in at least 2 of 200 de novo AML cases reported by The Cancer Genome Atlas Research Network,18 with the addition of 4 genes previously implicated in MDS/AML (JAK1, JAK2, JAK3, TYK2) and the exclusion of TTN and 3 pseudogenes (OR4H12P, LOC152845, LOC728843). Lastly, the human exome reagent (v3.0; Roche NimbleGen, Madison, WI) was used for exome capture. All sequence data has been deposited in the Database of Genotypes and Phenotypes under accession number phs000159.
Sequence alignment and variant calling
Illumina reads were aligned to the National Center for Biotechnology Information human genome assembly 37/hg19 reference sequence (GRCh37-lite) using Burrows-Wheeler Aligner v0.5.9.19 Binary alignment/map files were merged, and duplicates were marked using Picard v1.46 (http://picard.sourceforge.net). Five lanes yielded 12.2 Gb of aligned, deduplicated sequence. At least 90% of the target was covered ≥15× on average in all samples (supplemental Figure 2).
Single nucleotide variant (SNV) calls were generated from the union of 2 sets: (1) VarScan v2.2.620 (parameters: min-coverage, 3; min-var-frequation, 0.20; P =.10; strand-filter, 1; map-quality, 10) from SAMtools21 (r963) mpileup output (-q 10); and (2) SAMtools r963 filtered by snpFilter.pl using default parameters. Small insertion/deletion events (indels) were called using VarScan v2.2.6 using the same parameters. Both SNP and indel call sets were filtered to remove artifacts evident from misaligned reads, as previously described.20 Filter-passed variants were annotated with gene structure information using Ensembl release 67 transcripts. To identify possible copy number alterations, we computed the ratio of copy number change in each sample at each targeted exon, as follows: change = log2 × (sample_depth ÷ cohort_depth), where sample_depth is the average sequencing depth obtained for the individual for a given exon, and cohort_depth is the mean across all individuals.
Results
Detection of known pathogenic germ line variants
The proportion of families with predisposition to MDS/AML that can be explained by known genetic factors has not been clearly established. To address this question, we screened 12 known predisposition genes (supplemental Table 1) in 59 subjects from 17 families (supplemental Figure 3). SNVs or indels that met the following criteria were retained: coverage >20×, variant predicted to have translational consequences (including missense, nonsense, splice site, indel), variants segregated with affected individuals, and minor allele frequency <0.001 in population controls (dbSNP135, ESP650022,23). Seven SNVs fulfilling these criteria were identified in 7 individuals from 4 families.
Two known pathogenic variants in GATA2 (R398W, T354W)3,6,7 were identified in 2 families with previously unexplained familial MDS/AML (Table 1;24Figure 1A). The previously reported truncating RUNX1 variant (S388*) segregating in family 101514 was detected in both affected individuals and in 1 asymptomatic carrier who was 18 years of age at the time of sampling and clearly still at risk for development of MDS/AML (Table 1; Figure 1B).
These alleles were confirmed by Sanger sequencing and segregated with MDS/AML in the 3 affected families (supplemental Figure 4). The proband in family 1011, with clinical features suggesting Shwachman-Diamond syndrome, carried 2 predicted deleterious SBDS alleles, consistent with autosomal recessive inheritance in the offspring of a consanguineous marriage. The proband and her sister were also compound heterozygotes for 2 rare FANCA alleles, which are likely polymorphisms, because neither sister had stigmata of Fanconi anemia, and the alleles are not recognized as clinically significant (in ClinVAR or the Leiden Open-Source Variation Database 3.025,26). Furthermore, the unaffected sister carried 1 wild-type SBDS allele (rs113993993), suggesting that the proband’s phenotype was attributable to SBDS compound heterozygosity.
The proband of family 1001 (with clinical features suggesting the syndrome of monocytopenia with susceptibility to mycobacterial, fungal, and papillomavirus infection and myelodysplasia) was recently reported (subject 28.I.1) to carry a GATA2 variant in intron 5 (c.1017+572C>T) that disrupts a conserved ETS motif, resulting in haploinsufficient GATA2 expression.17 Sanger sequencing confirmed that this mutant allele was present in 3 of 4 affected individuals in this family (Table 1; supplemental Figure 4). One discordant result was obtained for an obligate carrier (004) who tested wild-type in a buccal sample that was obtained 9 years after a successful allogeneic stem cell transplant. The proband’s father (003) is heterozygous for the mutation but remains asymptomatic at age 57 years (Figure 1A). In total, a pathogenic germ line variant segregating with MDS/AML was identified in 5 of 17 (29%) families (Table 1).
Table 1
Pathogenic germ line variants detected by targeted sequencing

Figure 1
Familial MDS/AML pedigrees. (A) Partial pedigrees of families 1001 and 1002 with GATA2-associated MDS/AML; complete pedigrees are provided in supplemental Figure 1. Subjects who provided samples for sequencing are indicated by numerals. GATA2 genotypes are provided in Table 1. Two individuals with MDS acquired somatic ASXL1 mutations, as shown. (B) Partial pedigree of family 1015 with RUNX1-associated MDS/AML (left); complete pedigree is provided in supplemental Figure 1. The number of somatic variants (SNVs, indels) detected by exome sequencing in the 3 individuals (indicated by numerals in the pedigree) is shown in the circles (right). The size of the circles is proportional to the median VAFs of somatic SNVs in each case. NOS, not otherwise specified.
Detection of novel candidate germ line alleles
Because the known Mendelian MDS/AML predisposition genes are also targets of recurrent somatic mutation in sporadic cases of MDS/AML, we reasoned that novel inherited variants might be identified by screening a broader panel of RMGs in families with unexplained predisposition. To test this hypothesis, we screened normal samples from 39 individuals from 7 families (in whom the 12-gene panel screen failed to detect a pathogenic allele) for germ line variants in the 264-gene RMG panel (supplemental Figure 3). SNVs that met the following criteria were retained: coverage >20×, minor allele frequency <0.01 in population controls, and segregation with MDS/AML based on MendelScan analysis.27 We applied 2 additional conservative filters to produce a set of high-confidence germ line variants. First, we excluded SNVs with variant allele frequencies (VAFs) of >5% in unaffected individuals (to remove shared germ line variants that had passed a less-stringent VAF threshold of 20% in the initial call set).
Second, we excluded variants with VAFs <30% in affected cases (to remove false-positive sequence artifacts and somatic variants representing tumor contamination of the normal sample). An example of tumor contamination of the normal sample was a canonical DNMT3A mutation (R882H) present at a VAF of 16% in a buccal sample obtained from this individual (subject 1013-006) who had been in sustained remission for 6 years after induction and consolidation therapy for AML. Twenty-three variants in 16 genes met these criteria (supplemental Table 2). Four genes (HYDIN, MUC16, NMUR2, RNF213) warrant screening in additional families, because each had a novel allele that segregated with MDS/AML in >1 family.
Detection of somatic variants
The variable latency and penetrance of MDS/AML in carriers of known pathogenic inherited variants suggests that acquisition of cooperating somatic mutations is required for transformation. We first addressed the null hypothesis that RMGs in de novo MDS/AML are mutated at a similar frequency in familial cases. Paired tumor and normal samples from 15 familial MDS/AML cases (Table 2; supplemental Figure 3) were sequenced using the RMG capture reagent. Thirty-seven SNVs or indels were identified in 11 cases (no somatic SNVs/indels were identified in 4 cases), after removing calls failing the following stringent filters for somatic variants: <30 reads in the tumor or normal sample, calls with VAF >0 in normal or <10% in tumor, calls outside targeted exons, and calls failing manual review (supplemental Table 3). The median number of somatic mutations in these genes was significantly lower in familial cases compared to de novo AML cases18 (Figure 2A), which refuted the null hypothesis.
Table 2
Characteristics of patient cohort selected for tumor/normal sequencing

Figure 2
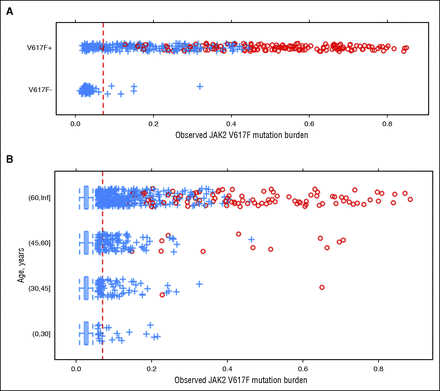
Somatic variants in familial MDS/AML vs de novo AML. (A) Targeted sequencing of known RMGs demonstrated fewer mutations in familial cases compared to de novo cases (median, 2.0 vs 5.0; P =.0013 by 2-tailed Mann-Whitney). (B) Somatic mutation VAFs detected by exome sequencing in asymptomatic RUNX1 carriers and familial MDS and AML cases from GATA2 or RUNX1 families are shown. Age at sample collection is indicated by the x-axis labels (black, RUNX1 carrier; red, GATA2 carrier). Clonal hematopoiesis was detectable in 6 of 9 asymptomatic RUNX1 carriers.
To determine whether the lower frequency of RMG mutations in familial MDS/AML cases simply reflected a lower burden of total somatic mutations compared to de novo cases, we performed exome sequencing on paired tumor and matched normal samples from 19 subjects (Table 2). We focused exclusively on families with known pathogenic RUNX1 or GATA2 germ line alleles to reduce the likelihood that some cases were sporadic (supplemental Figure 3). In all cases, the pathogenic germ line allele was detected in sequence data from the normal sample, except for the intronic GATA2 variant in family 1001 (which was not captured by the exome reagent). One hundred eighty-three SNVs/indels were identified in 16 subjects (no somatic SNVs/indels were detected in 3 asymptomatic carriers; supplemental Table 4).
Excluding the asymptomatic RUNX1 carriers, the median number of somatic SNVs/indels per case was identical in familial and de novo AML cases (n = 13 for both). This finding suggests that the lower frequency of RMG mutations in familial cases may not be explained by a lower of frequency of mutations overall, but rather by a different landscape of RMGs in these cases. Consistent with this interpretation, local realignment and manual review of the exome data failed to identify insertions at NPM1 or FLT3 in any of the familial AML cases, in striking contrast to their high mutation frequency in de novo AML.
Although the current study was not powered to detect novel recurrent somatic mutations in familial MDS/AML, frameshift mutations in PDS5B, which encodes a cohesin-associated factor, were identified in 2 cases (1 each in GATA2 and RUNX1 families). Recurrent frameshift mutations in PDS5B have been reported in gastric, colorectal, and breast cancer.28,29 Combining results from exome and targeted sequencing, somatic mutations in the following well-characterized MDS/AML–associated genes were detected in 2 or more familial cases: BCOR, DNMT3A, ASXL1, PTPN11, and STAG2 (supplemental Tables 3 and 4).
Somatic mutations in ASXL1 have been reported by several groups in association with germ line GATA2-associated MDS/AML.30,31 Consistent with these reports, we detected canonical loss-of-function ASXL1 mutations in 2 of 4 MDS/AML cases from GATA2 families that were subjected to exome sequencing (Figure 1A). Recently, somatic mutations in CDC25C were reported in 7 of 13 (53%) patients with hematologic malignancies from FPDMM families.32 The reported mutations were all missense substitutions at codons 233, 234, 344, or 437. In contrast to this report, we detected no somatic mutations in CDC25C by exome sequencing in 13 individuals from FPDMM families. We also performed Sanger sequencing using amplicons containing these 4 codons and confirmed that somatic mutations in CDC25C were absent in these 13 cases and in asymptomatic carriers from FPD/AML families.
The median VAFs of SNVs from the MDS and AML samples were similar (ranging from 23.5% to 63.6% and from 16.4% to 53.6%, respectively; Figure 2B), consistent with results from whole-genome sequencing, suggesting that the extent of clonal hematopoiesis is similar in MDS and AML.33 Strikingly, a majority of the samples from asymptomatic RUNX1 carriers that were tested (67%) harbored detectable somatic mutations, with median VAFs in the same range as seen in the MDS/AML samples (20.5%-71.4%), including in 1 case a canonical DNMT3A variant, suggesting that clonally skewed hematopoiesis frequently precedes development of overt MDS/AML in these carriers (Figure 2B). Several recent studies have detected clonally skewed hematopoiesis in asymptomatic elderly individuals that is often associated with mutations in epigenetic regulators.34-36 However, clonal hematopoiesis was rarely (<1%) present in individuals under the age of 50 years in these reports, whereas all 6 of the asymptomatic RUNX1 carriers with clonal hematopoiesis in this study were under the age of 50 years at the time of sample collection. Although the current sample size is small, the cumulative risk of developing clonal hematopoiesis reached 81% by age 50 years in RUNX1 carriers. Further study is required to determine whether this biomarker could be used to prospectively identify RUNX1 carriers at higher risk for evolution to MDS/AML (Figure 3).
Clonal evolution in RUNX1 carriers. Asymptomatic carriers of pathogenic germ line RUNX1 variants develop early-onset clonal hematopoiesis (cumulative risk of 81% by age 50 years). As depicted in the model, this finding provides a rationale for testing the hypothesis that clonal hematopoiesis may provide a biomarker for early detection of disease progression in high-risk families.
Discussion
Familial aggregation of MDS and AML occurs more frequently than expected by chance, suggesting that inherited factors influence susceptibility to these diseases.37 In contrast to the major cancer predisposition syndromes, an inherited basis is frequently not considered, and a genetic basis is rarely sought when cases of MDS/AML co-occur in biological relatives. This may be due in part to limited familiarity with these syndromes, the expensive and time-consuming process of serial single-gene testing in clinical laboratories, and the lack of commercially available tests in some cases. Next-generation sequencing-based gene panel testing may provide a more cost-effective and efficient strategy to identify clinically significant variants in high-risk families. Using a small targeted sequencing panel, we identified a pathogenic germ line variant in 5 of 17 (29%) MDS/AML pedigrees, a frequency similar to the proportion of high-risk breast cancer families that can be explained by known genes.38 Because this panel omitted several genes recently implicated in MDS/AML susceptibility (eg, ANKRD26, SRP72, DDX41, ETV6), this frequency is likely an underestimate.
GATA2 variants were the most frequently identified explanation for familial MDS/AML in this cohort (3/5 families). Of importance, none of the targeted sequencing approaches used here adequately assessed noncoding regions of the GATA2 locus that are known to be associated with familial MDS/AML, implying that the true frequency of GATA2-associated disease may be higher. A recent study of 71 individuals with idiopathic MDS or bone marrow failure (half of whom lacked a family history) found a similarly high proportion of GATA2 variants (5/8 individuals in whom a causal variant could be identified).39
Recognition of familial MDS/AML and identification of its genetic basis are important for several reasons. Siblings who may be asymptomatic carriers must be recognized and should be excluded as donors for allogeneic stem cell transplantation. Carriers should be followed by hematologists for surveillance and early detection of clonal progression and management of associated bleeding disorders and other organ dysfunction. Families should be referred to genetic counselors for discussions of reproductive risk. Because a genetic explanation can currently be provided in a minority of cases, additional research is required to identify all of the genes associated with susceptibility, and the identification of these genes may provide new insight into AML pathogenesis. Several novel candidates identified here require testing for replication in additional families. Unbiased sequencing is likely to identify additional candidates in coding (and potentially noncoding) regions of the genome. In some instances, co-occurrence of MDS/AML within biological relatives may be due to chance or shared environmental exposure, rather than to inherited genetic factors.
The identification of recurrent somatic alterations in these cases may provide targets for novel therapy and early detection. We found that the total number of mutations in protein-coding genes per case is similar in familial MDS/AML and de novo AML, but the spectrum of recurrently mutated genes appears to be different. We identified recurrent frameshift mutations in PDS5B, adding to the growing body of literature on the importance of the cohesin complex in the pathogenesis of myeloid leukemias.40,41 We confirmed the previously reported co-occurrence of somatic mutations in ASXL1 with germ line mutations in GATA2,6,30 but could not confirm a recent report of frequent somatic mutations in CDC25C in RUNX1 carriers,32 perhaps because of ethnic differences between patient cohorts or other factors.
The recent recognition that asymptomatic clonal hematopoiesis occurs with increasing frequency as a function of age has raised questions about the natural history and clinical significance of this phenomenon.42 Although the presence of clonal hematopoiesis has been associated with elevated risk of leukemic transformation, the absolute risk appears to be small.35,36 We detected clonal hematopoiesis in 67% of asymptomatic RUNX1 carriers under the age of 50 years, representing a cumulative risk of 81% by age 50 years. Although these results are preliminary, this rate is several orders of magnitude higher than what has been reported for this age group in the general population. Serial analysis of clonal hematopoiesis warrants further study to determine whether it may provide a biomarker for early detection of disease progression in mutation carriers from these high-risk families.
Authorship
Contribution: T.J.L., J.E.C., L.A.G., and T.A.G. designed the study; J.E.C., K.P., I.P., G.L.U., E.M.B., M.L., J.R., K.E., S.H., and L.A.G. obtained the samples and constructed the pedigree; K.-L.K., J.S., D.K., C.A.M., D.S., R.F., M.O., C.F., A.J., P.W., J.F.D., D.C.L., M.J.W., J.W., R.W., and T.A.G. produced and analyzed the data; and J.E.C., T.J.L., L.A.G., and T.A.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy A. Graubert, Massachusetts General Hospital Cancer Center, 10 North Grove St, Lawrence House 204, Boston, MA 02114; e-mail: tgraubert{at}partners.org.
Acknowledgments
The authors wish to acknowledge the individuals and families who participated and remain engaged in the research.
This work was supported by National Institutes of Health National Cancer Institute grants CA101937 (T.J.L.), CA157439 (J.R.), and K12 CA139160 (J.E.C.); the Cancer Research Foundation (L.A.G., J.E.C.); and the Children’s Research Fund (J.R.).